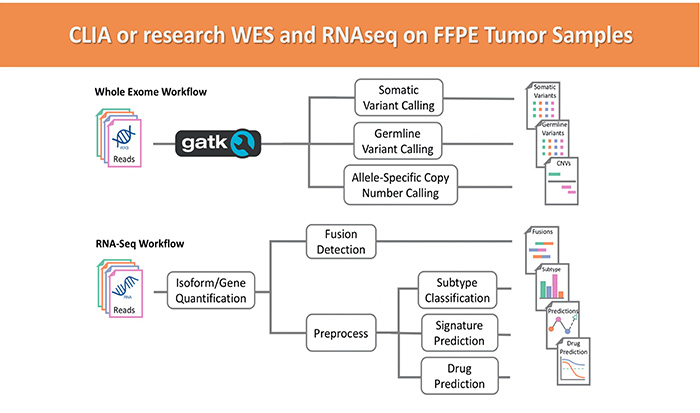
We offer RNAseq and whole exome sequencing (WES). Both are powerful tools for measuring gene expression changes and mutational burden, respectively, with many applications including diagnostic and prognostic research and clinical applications. WES sequencing refers to genomic DNA sequencing that is enriched for exonic regions. RNA sequencing, on the other hand, may or may not be enriched (for example for polyadenylated transcripts). Our expert scientists are available to consult with you on project-specific details and appropriate experimental design.
RNA-Seq is a powerful tool for measuring gene expression changes with many applications including:
- Assessing treatment response as a predictive biomarker at baseline or over the course of treatment (in response to standard of care treatment, in response to immune treatment and longitudinal biopsies).
- Primary- early and late stage and metastatic disease types
- Predictive biomarkers for risk/therapeutic stratification
- Identification of response vs resistance to treatment
Methodology:
We offer comprehensive service which includes:
- Isolation of total RNA from cells or tissues
- Assessment of RNA quality/integrity
- polyA+ RNA enrichment or rRNA depletion
- Library generation and QC of NGS library
- Next-generation sequencing using the Illumina platform
- Comprehensive bioinformatics and data analysis
We work with many different FFPE embedded tissue types (with tumor micro-dissection when applicable) and cell suspensions. Our specimens are always certified-pathologist reviewed and annotated. We implement well standardized and highly sophisticated analysis to best interrogate your data from a clinically relevant perspective. Both NGS based services are available in research-only and CLIA/CAP certified environments always following GLP guidelines.
Specimen Requirements:
The NGS analysis is compatible with FFPE tissue. Sample requirements and preparation guidelines are available upon request. IMCO@ohsu.edu
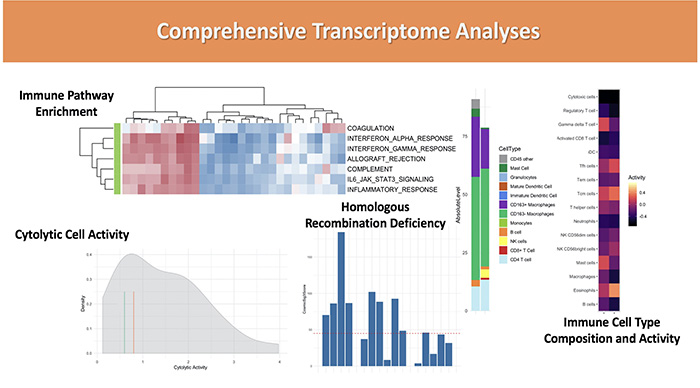
Data visualization and interpretation examples: