The nanoString® GeoMx platform supports quantitative analysis of proteins, phospho-proteins and mRNAs from user defined cell populations across sections of FFPE tumor tissue. The Knight Cancer Institute is a designated Center of Excellence for GeoMx applications and is working closely with nanoString in the development and validation of new biomarker targets. Over 100 proteins and 1,800 mRNAs can be rapidly analyzed with this technology.
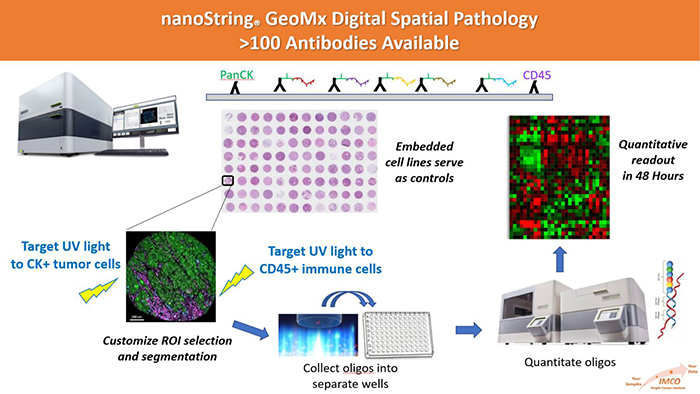
Most approaches to multiplexed analysis of biomarkers in FFPE tissue depend on an analog readout (immunofluorescence or histochemical stain) and are limited to a few dozen targets. The GeoMx platform uses oligonucleotide-tagged antibodies and probes that provide a digital readout with a very high dynamic range. Highly focused UV light is used to release the tags from the cells of interest (tumor, immune cells, stromal cells, etc.) and these are quantitated with either the nCounter system or next-gen sequencing. Additional information can be found at:
GeoMx® Digital Spatial Profiler
mRNA analysis:
GeoMx® Cancer Transcriptome Atlas
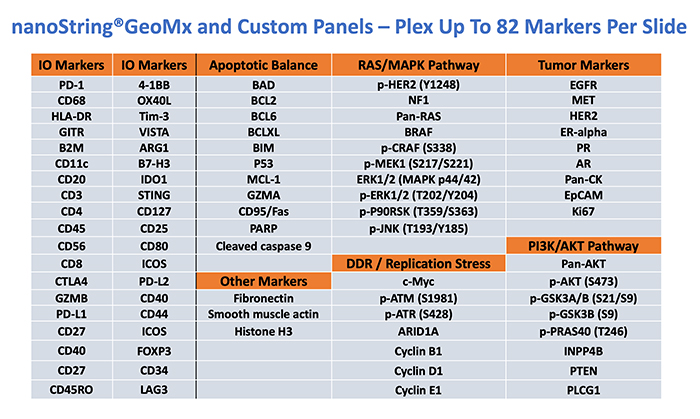
The GeoMx® platform can serve a wide spectrum of clinical research applications. Combinations of the markers listed below can be put together to provide a customized approach for every project. Find out more by contacting IMCO@ohsu.edu
Available Protein markers
nanoString® GeoMx Assays
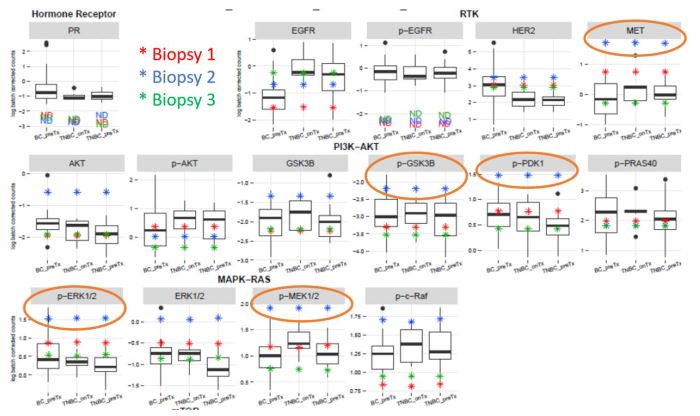
Sample report
Analytical Validation of Quantitative Intracellular Protein poster presented at AMP